20 Sep New papers from current and former Lister Fellows offer insights into antimicrobial resistance and DNA repair
This week, we review two papers published in Nature and Nature Communications by leading researchers Professor Ross Chapman, based at the Weatherall Institute of Molecular Medicine, and Professor Serge Mostowy, based at The London School of Hygiene & Tropical Medicine.
Serge Mostowy’s latest paper reveals potential new treatment for bacterial infections

In a paper published in Nature Communications, Professor Serge Mostowy and his team describe details of the way in which proteins called septins identify and interact with bacteria. This paves the way towards using septins as a targeted treatment for specific bacterial infections and in combatting antimicrobial resistance.
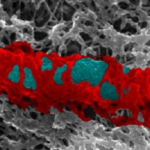
The research follows on from the Mostowy Lab’s previous work, which outlined the potential of septins to combat antimicrobial resistance. The team discovered that host cells create septin cages to restrict the movement and growth of Shigella bacteria and flags them for destruction by the host’s immune system.
The new research – led by first author Damián Lobato-Márquez – delves deeper into the molecular mechanisms of this process. It used a ‘bottom-up’ approach to synthesize septin cages in vitro.
“This paper proposes a novel approach to future medicine, with the potential for great impact in the fight against antimicrobial resistance,” says Serge. “It reports a milestone discovery in septin cage biology and cell-autonomous immunity. As such, our findings should encourage further work to exploit the septin cytoskeleton in recognising and treating bacterial infection.”
Serge and his colleagues found that septins can distinguish between different species of bacteria, behaving differently towards Escherichia coli, Shigella, and Mycobacteria. The proteins alter their behaviour according to whether or not bacteria are dividing, the shape of their cell, and the makeup of the bacterial surface.
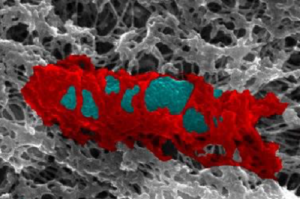
Proteins build septin cages (red) to trap bacteria (blue).
The in vitro study showed that septins independently and selectively bind to dividing Shigella. They appear to bind to the surface of Shigella cells, exposed when bacteria produce IcsA protein, and that the bacteria attempt to prevent this binding and septin cage entrapment by producing lipopolysaccharide as a physical barrier on their surface. Unexpectedly, it was observed that septins bind more quickly and more easily to the surface of Mycobacteria.
The team also made the fascinating discovery that septins can sense the curvature of the poles of Shigella cells and then bind to these regions – meaning that septins can identify bacteria cells from their shape.
You can read the full paper, “Mechanistic insight into bacterial entrapment by septin cage reconstitution”, on the Nature Communications website.
“Next, it is of great interest to identify the full breadth of septin targets on the surface of different bacteria that threaten human health,” says Serge. “A better understanding of septin interactions with the bacterial surface will lead to innovative applications in the clinic.”
Professor Serge Mostowy is a professor at The London School of Hygiene & Tropical Medicine, a globally renowned research and education institution with a substantial track record in public health. You can find out more about his research work on his laboratory’s website.
Ross Chapman’s new research describes mechanisms behind the repair of cancer-causing mutations

As part on their ongoing mission to document and understand DNA repair, Professor Ross Chapman’s lab have revealed the precise mechanism behind how the BRCA1 protein detects and engages with DNA breaks in the genome to help prevent breast and ovarian cancers developing.
People with mutations in BRCA1 are at much higher risk of developing breast and ovarian cancer. This is because BRCA1 plays a key role in accurately coordinating the repair of DNA breaks in dividing cells. But until now it has been unclear how BRCA1 functions and why its loss can lead to alternative, more inaccurate repair mechanisms that speed up tumour development.
In their latest paper, current Lister Fellow Ross and his team describe that BRCA1 works alongside the partner protein BARD1, which recognises a ‘histone code’ generated in the chromatin that packages our DNA.
“Our work shows that BRCA1’s partner BARD1 contains two specialised domains that function as a unit, enabling it to sense two important signals in the chromatin packaging of of our genomes.” says Ross.
“One of these says there is a DNA break close by, while the other signal indicates that this region of the genome has recently been replicated. By integrating these two signals, BRCA1 and BARD1 can assemble the cells DNA repair machinery at sites of DNA damage, where they ensure that the intact copy of the damaged DNA strand is used as a template for error-free DNA repair. This is vital, as DNA breaks arise frequently during cell division, and we know that their inaccurate repair in cells lacking BRCA1 gives rise to cancer causing mutations.”
The paper, “BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination”, was published in Nature in July 2021.
The research also showed the histone code used by BARD1 was closely related to the signal recognised by 53BP1. This DNA repair protein generates purposeful and important recombination events within antibody genes, which allows our immune systems to generate a variety of antibodies with differing antigen and tissue specificities.
When 53BP1 was deleted, BRCA1 stopped relying on BARD1 to repair DNA. This suggests that BRCA1 evolved to counteract the threat that this mutagenic DNA repair would otherwise pose to the stability of our genomes, if it was allowed to access spontaneously occurring DNA breaks during cell division.
Professor Ross Chapman is based at the Radcliffe Department of Medicine at the Weatherall Institute of Molecular Medicine. You can find out more about his research into the role of recombination mechanisms in cancer and immunity on his laboratory’s webpage.
